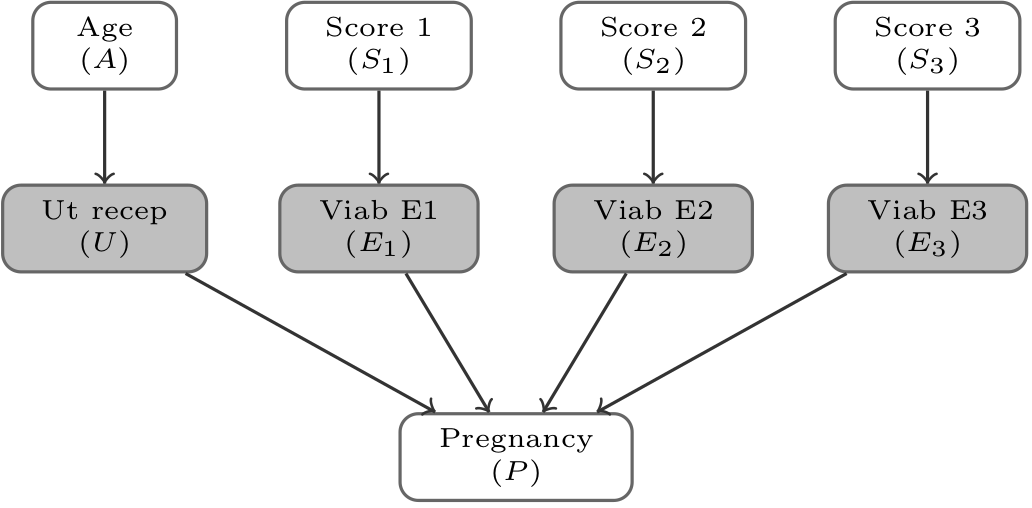
data {
int N;
array[N] int A;
array[N, 3] int S;
array[N] int P;
}
parameters {
simplex[3] pA;
simplex[4] pS;
vector<lower=0, upper = 1>[3] pU;
vector<lower=0, upper = 1>[3] pE_;
}
transformed parameters {
vector<lower=0, upper = 1>[4] pE;
pE[1] = 0;
pE[2:4] = pE_;
}
model {
// Priors
pA ~ dirichlet(rep_vector(1.0/3, 3));
pS ~ dirichlet(rep_vector(.25, 4));
pU ~ beta(1,1);
pE_ ~ beta(1,1);
// Likelihood
for (n in 1:N) {
target += categorical_lpmf(A[n] | pA);
target += categorical_lpmf(S[n,] | pS);
}
for (n in 1:N) {
if (P[n] > 0) {
target += log(pU[A[n]]);
if (P[n] == 1) {
target += log(
pE[S[n,1]] * (1 - pE[S[n,2]]) * (1 - pE[S[n,3]]) +
(1 - pE[S[n,1]]) * pE[S[n,2]] * (1 - pE[S[n,3]]) +
(1 - pE[S[n,1]]) * (1 - pE[S[n,2]]) * pE[S[n,3]]
);
}
if (P[n] == 2) {
target += log(
pE[S[n,1]] * pE[S[n,2]] * (1 - pE[S[n,3]]) +
(1 - pE[S[n,1]]) * pE[S[n,2]] * pE[S[n,3]] +
pE[S[n,1]] * (1 - pE[S[n,2]]) * pE[S[n,3]]
);
}
if (P[n] == 3) {
target += log(pE[S[n,1]] * pE[S[n,2]] * pE[S[n,3]]);
}
} else {
target += log(
1 - pU[A[n]] * (
pE[S[n,1]] * (1 - pE[S[n,2]]) * (1 - pE[S[n,3]]) +
(1 - pE[S[n,1]]) * pE[S[n,2]] * (1 - pE[S[n,3]]) +
(1 - pE[S[n,1]]) * (1 - pE[S[n,2]]) * pE[S[n,3]] +
pE[S[n,1]] * pE[S[n,2]] * (1 - pE[S[n,3]]) +
(1 - pE[S[n,1]]) * pE[S[n,2]] * pE[S[n,3]] +
pE[S[n,1]] * (1 - pE[S[n,2]]) * pE[S[n,3]] +
pE[S[n,1]] * pE[S[n,2]] * pE[S[n,3]]
)
);
}
}
}
generated quantities {
array[N] vector[4] ppm;
vector[N] yrep;
for (n in 1:N) {
ppm[n, 1] = 1 - pU[A[n]] * (
pE[S[n,1]] * (1 - pE[S[n,2]]) * (1 - pE[S[n,3]]) +
(1 - pE[S[n,1]]) * pE[S[n,2]] * (1 - pE[S[n,3]]) +
(1 - pE[S[n,1]]) * (1 - pE[S[n,2]]) * pE[S[n,3]] +
pE[S[n,1]] * pE[S[n,2]] * (1 - pE[S[n,3]]) +
(1 - pE[S[n,1]]) * pE[S[n,2]] * pE[S[n,3]] +
pE[S[n,1]] * (1 - pE[S[n,2]]) * pE[S[n,3]] +
pE[S[n,1]] * pE[S[n,2]] * pE[S[n,3]]
);
ppm[n, 2] = pU[A[n]] * (
pE[S[n,1]] * (1 - pE[S[n,2]]) * (1 - pE[S[n,3]]) +
(1 - pE[S[n,1]]) * pE[S[n,2]] * (1 - pE[S[n,3]]) +
(1 - pE[S[n,1]]) * (1 - pE[S[n,2]]) * pE[S[n,3]]
);
ppm[n, 3] = pU[A[n]] * (
pE[S[n,1]] * pE[S[n,2]] * (1 - pE[S[n,3]]) +
(1 - pE[S[n,1]]) * pE[S[n,2]] * pE[S[n,3]] +
pE[S[n,1]] * (1 - pE[S[n,2]]) * pE[S[n,3]]
);
ppm[n, 4] = pU[A[n]] * (pE[S[n,1]] * pE[S[n,2]] * pE[S[n,3]]);
yrep[n] = categorical_rng(ppm[n, ]);
}
}